What Are Asparaginases?
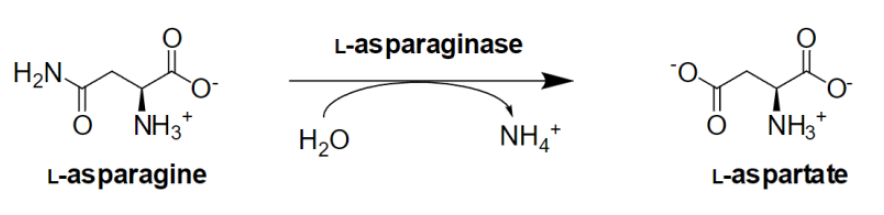
l‑Asparaginases are a group of enzymes that catalyse the hydrolysis of l‑asparagine to l‑aspartate and ammonium, a reaction important in nitrogen metabolism and maintaining cellular and physiological homeostasis. This class of catalytic proteins is found in archaea, prokaryotes, and eukaryotes, including humans, and sequences are commonly present in several isoforms. Consequently, it emerges as a highly diverse group, characterized by various quaternary structures with distinct amino acid sequences, while displaying a wide spectrum of affinities not only for l‑asparagine but often also for other substrates. Click the reaction scheme for more details.
Medical Applications
l‑Asparaginases play a pivotal role in treating acute lymphoblastic leukemia and rank among the most successful biopharmaceuticals, having served as effective anticancer agents since 1978. They function by efficiently depleting l‑asparagine from the bloodstream, which is an essential nutrient that many cancer cells cannot produce independently, while healthy cells retain the ability to synthesize their own l‑asparagine.
Food Industry Applications
In the food industry, l‑asparaginases are employed in reducing the formation of acrylamide, a neurotoxic compound that develops when l‑asparagine reacts during the heat processing of carbohydrate-rich foods. By breaking down l‑asparagine before thermal processing, these enzymes effectively minimize acrylamide levels in processed food products, thereby reducing potential health risks associated with this toxic substance.
Current Challenges
Despite their success in cancer therapy and industry applications, current l‑asparaginase preparations face several significant challenges, including pronounced immunogenicity, limited stability, and suboptimal activity or substrate affinity.
Enzyme Diversity
l‑Asparaginases are a diverse group of enzymes, and understanding their natural diversity and structure-activity relationships is essential for addressing current limitations. Related enzymes include l‑glutaminases, mixed asparaginases/glutaminases, β-aspartyl-peptidases, glutamyl-tRNAGln amidotransferases, and aspartylglucosaminidases.
About the Database
The Asparaginase Database is a resource that provides organized l‑asparaginase sequences and structures within their phylogenetic families along with biochemical annotations for characterized enzymes. The database is built on a comprehensive phylogenetically driven l‑asparaginase classification that integrates existing biochemical knowledge with expanding sequence data. This system is not only free of historical ambiguities but could also aid scientists with understanding and discovering l‑asparaginases with improved functional or structural properties.
The current release covers a manually curated set of 101 experimentally studied proteins from the literature, a manually curated set of 127 sequences from SwissProt and 126,085 automatically classified sequences.
The Classification
The l‑asparaginase classification that underpins the database is based on extensive sequence (and structure) analyses of known and predicted l‑asparaginases. As these enzymes form a complex and intertwined group, sequence similarity analysis was selected as the best primary driver of the classification, rather than other factors like taxonomy, cellular location or enzyme kinetics, enabling a more objective and natural classification less biased by the order of scientific discovery. This bioinformatics-centred approach also makes the classification more robust to future discoveries.
Asparaginase Classes
There are three fundamental classes of l‑asparaginases, each with unique conserved motifs and structures. No homology could be inferred between these classes of l‑asparaginases, as they show very low mutual sequence identity. Sequences are categorized into Class 1 (historically bacterial-type), Class 2 (plant type) and Class 3 (R. etli-type) l‑asparaginases. Sequences in these classes were clustered and aligned independently. This allowed further division into new phylogenetic clans and families inferred from a comprehensive phylogenetic analysis.


